Part:BBa_K4278717
l-tef1-atf1-cyc1-tefp-nrsr-teft-r
l-tef1-atf1-cyc1-tefp-nrsr-teft-r
Profile
Name: l-tef1-atf1-cyc1-tefp-nrsr-teft-r
Base Pairs: 4450 bp
Origin: Saccharomyces cerevisiae genome and pHCas9-Nours plasmid
Properties: an ATF1 and NrsR gene reading cassette
Usage and Biology
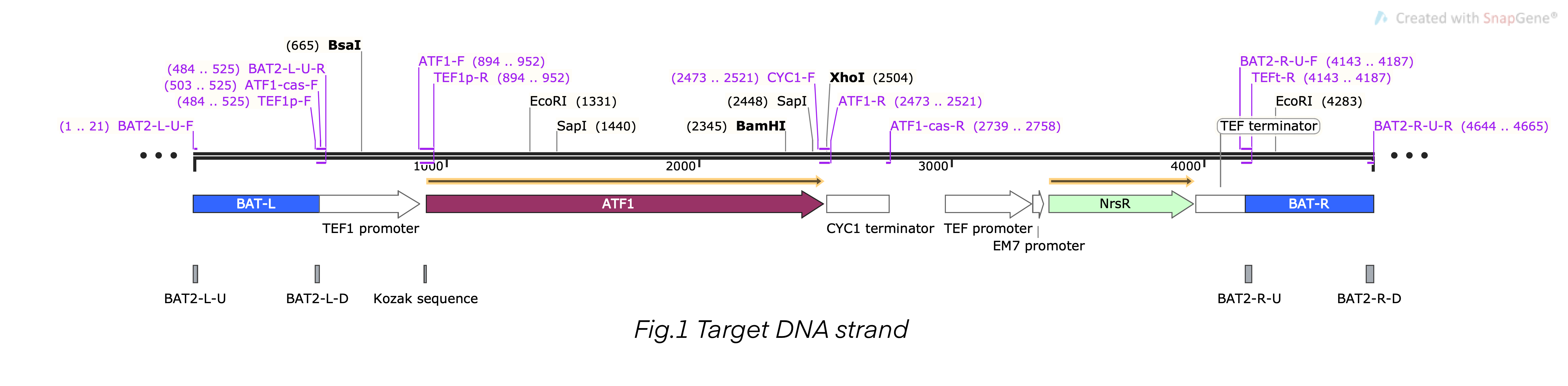
This composite part is the recombinant DNA fragment constructed by TEF1 promoter (BBa_K4278701), ATF1 DNA fragment (BBa_K4278702), CYC1 terminator (BBa_K4278703), TEF promoter (BBa_K4278705), NrsR DNA fragment (BBa_K4278706), and TEF terminator (BBa_K4278707). This DNA fragment also contained a homologous arm designed according to BAT2, we fused the 503bp upstream sequence of BAT2 (BBa_K4278700) at the 5’-terminal of the composite DNA sequence, and we fused the 501bp downstream sequence of BAT2 (BBa_K4278708) at the 3’-terminal. The homologous arms were used to replace the BAT2 gene fragment in the S. cerevisiae genome in a recombinant way [1-6].
Experimental approach
1. Construct the composite DNA fragment
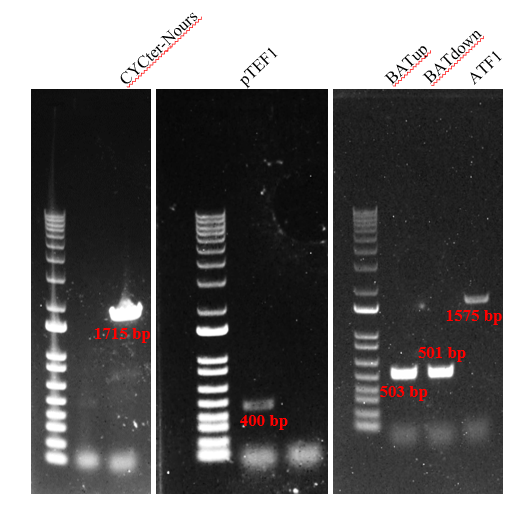
Firstly, we amplified five fragments, which were CYC1-TEFp-NrsR-TEFt, pTEF1, BATup, BATdown, and ATF1. The results (Figure 2) indicated that the five target DNA strands are successfully amplified.
Secondly, we fused the DNA fragments in figure2 by PCR. The following result of the gel electrophoresis shows the gene strands are successfully amplified (Figure 3).
Proof of function
1. Measure the growth curve of ATF1 transformants
The constructed DNA fragment (BBa_K4278717) was transformed into the S. cerevisiae and replaced the BAT2 gene in the genome through a recombinant way and we named the transformants SF-1. In order to measure if the integrated gene interferon the growth of the host strain, we measured the growth curve arranged 32h and compared it with the wildtype AQ yeast. This result indicates that the genetically modified Saccharomyces cerevisiae SF-1 that we aimed to reduce drunk phenomenon caused no harmful effects on the survival and growth of the organism itself. Therefore, giving support for applications in the wine fermentation industry (Table 1).
2. Functional test
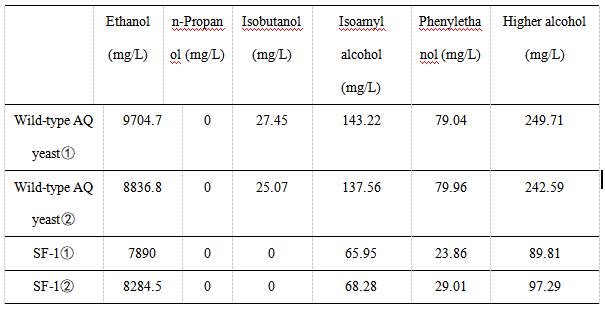
We use gas chromatography to measure the content of higher alcohols. The following table shows the experimental data of various alcohols produced by wild-type AQ yeast and SF-1 yeast. We did two sets of experiments for each yeast. The experimental data showed that the ethanol content of SF-1 yeast was similar to that of wild-type AQ yeast, indicating that the knockout of the BAT2 gene and integration of the ATF1 gene had little effect on the production of ethanol.
As indicated in Table 2, the content of high alcohol produced by AQ yeast , AQ yeast , Fragments F, Fragments F, Plasmid pYES2-ATF1 , and Plasmid pYES2-ATF1 is 249.71, 242.59, 89.81, 97.29 mg/L, respectively.
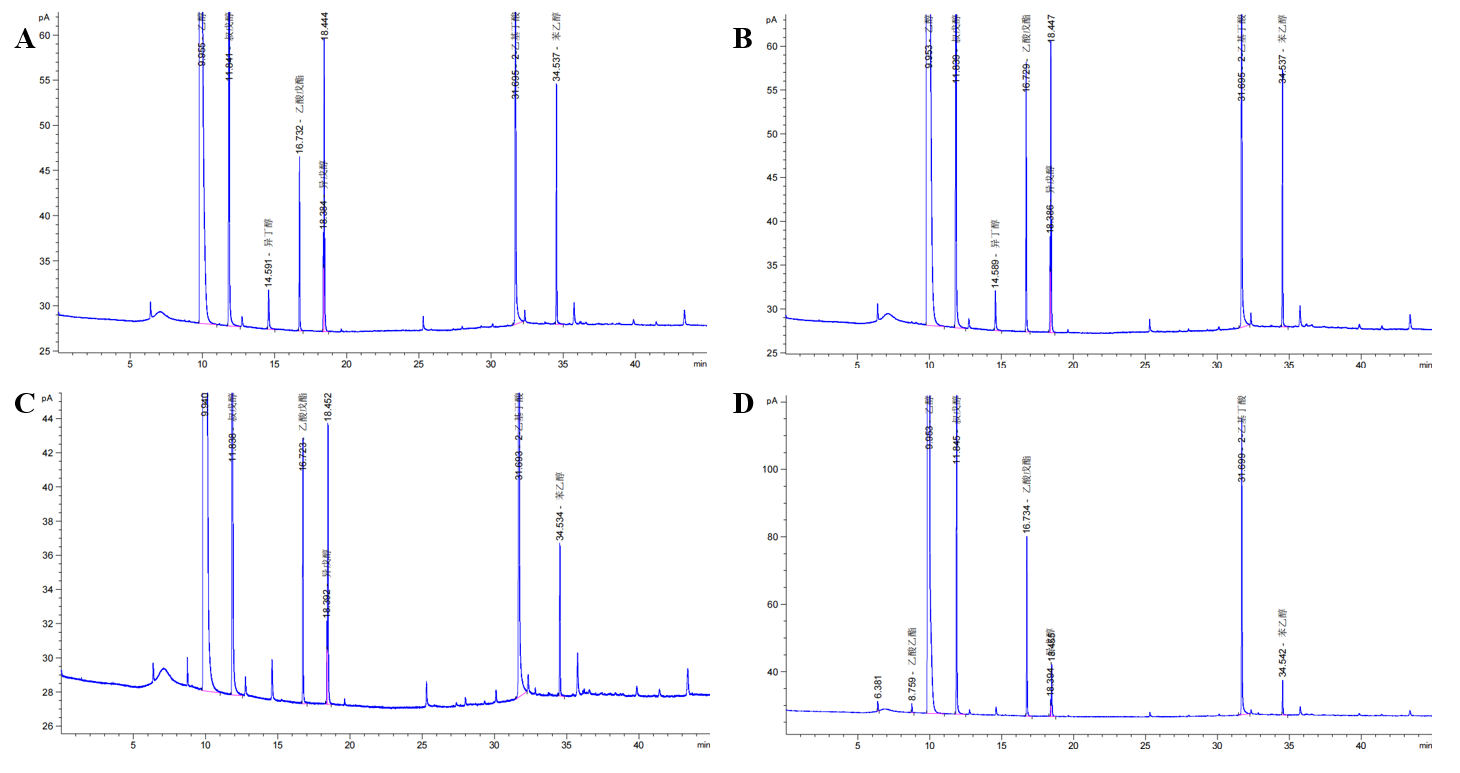
In Table 2 and Figure 4, we can find that the percentage of higher alcohols produced by wild-type yeast was significantly higher than that produced by SF-1. These results indicated that replacing the BAT2 gene with the ATF1 gene of alcohol acetyltransferase could reduce the production of higher alcohols in yeast strains. In other words, the wine produced by SF-1 engineering bacteria has a low content of higher alcohols, which is not likely to cause symptoms such as headache and thirst after drinking. Our study provides solutions to problems such as hangovers and headaches after drinking alcohol, which have certain practical significance.
Improvement of an existing part
Our composite part BBa_K4278717 is composited by the promoters, ATF1 gene fragment, NrsR gene fragment, and terminators. We amplified the DNA fragment and transformed it into S. cerevisiae to replace the BAT2 DNA fragment in the strain genome. It is improved based on the existing part BBa_J45014. In 2006, group iGEM2006_MIT designed a basic part BBa_J45014, ATF1, but they only provided the DNA sequence of it. To control the content of higher alcohols during fermentation, by reading literature and consulting experts in related fields, we replaced the BAT2 gene with the ATF1 reading cassette in the host strain. By detecting the fermentation product of the transformants, it was further confirmed that the engineered S. cerevisiae has reduced the production of higher alcohols which could be used to improve the quality of the wine in factories in the future [7].
References
1.Zhang J., Zhang C., Wang J., Dai L., Xiao D. (2014) Expression of the Gene Lg-ATF1 Encoding Alcohol Acetyltransferases from Brewery Lager Yeast in Chinese Rice Wine Yeast. In: Zhang TC., Ouyang P., Kaplan S., Skarnes B. (eds) Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012). Lecture Notes in Electrical Engineering, vol 249. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-37916-1_5.
2.Ma L, Huang S, Du L, Tang P, Xiao D. Reduced Production of Higher Alcohols by Saccharomyces cerevisiae in Red Wine Fermentation by Simultaneously Overexpressing BAT1 and Deleting BAT2. J Agric Food Chem. 2017 Aug 16;65(32):6936-6942. doi: 10.1021/acs.jafc.7b01974.
3.Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res. 2006 Aug;6(5):726-43. doi: 10.1111/j.1567-1364.2006.00057.x.
4.Dai L., Zhang C., Zhang J., Qi Y., Xiao D. (2014) Effects of IAH1 Gene Deletion on the Profiles of Chinese Yellow Rice Wine. In: Zhang TC., Ouyang P., Kaplan S., Skarnes B. (eds) Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012). Lecture Notes in Electrical Engineering, vol 249. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-37916-1_42.
5.Choi YJ, Lee J, Jang YS, Lee SY. Metabolic engineering of microorganisms for the production of higher alcohols. mBio. 2014 Sep 2;5(5):e01524-14. doi: 10.1128/mBio.01524-14.
6.孙中贯,刘琳,王亚平,王雪山,肖冬光.酿酒酵母高级醇代谢研究进展[J].生物工程学报,2021,37(2):429~447.
7.于洪梅. 气相色谱法分析啤酒中 5 种高级醇的方法研究[J]. 食品研究与开发, 2018, 39(18):5
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1013
Illegal BglII site found at 4307 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 3473
Illegal AgeI site found at 3191 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 679
Illegal SapI.rc site found at 1440
| None |